On the lookout for viruses
PCR and rapid tests are the most common methods of virus detection. Whereas PCR analyzes a DNA sample, rapid tests detect antigens, i.e., substances foreign to the body, and antibodies, proteins produced by the body to counter antigens.
However, each method has its drawbacks. With PCR, it’s time – it takes four hours to produce the necessary amount of DNA and complete the test. Rapid tests offer quicker results but have an 11-48% false positive rate (depending on the symptoms). Moreover, both of these methods aim to detect a specific virus supposedly present in the patient’s system.
A research team from ITMO University and Smorodintsev Research Institute of Influenza have suggested a multipurpose detection method based on surface-enhanced Raman spectroscopy (SERS) and machine learning. It can rapidly detect coronavirus (hCoV-19/Russia/St.Petersburg-3524/2020) and influenza type A (A/California/07/2009, A(H1N1)pdm09) and B (B/Hong Kong/269/2017) viruses in saliva or throat swabs.

Placing a sample onto a SERS surface. Photo by Dmitry Grigoryev / ITMO.NEWS
How it works
Every virus has a capsid, a shell with proteins specific for every virus. Using these proteins, viruses connect to cells, enter them, and then start copying and producing more viral particles. As any proteins, these consist of amino acids; they are synthesized based on the genetic material of the virus. Different amino acids have different reactions to light – they can absorb or emit radiation. Depending on the amino acids that make up a protein, it will react differently to laser radiation. The light from the laser is diffused in the sample, presenting a spectrum. If we know the Raman spectrum of a substance, including a virus, we can identify it.
SERS, the approach used by ITMO scientists, allowed them to trace the changes in a sample’s spectrum caused by viral surface proteins. This method relies on metal nanoparticles as amplifiers: when they interact with laser radiation, they create hot spots, local maximums. If a virus connects to such a spot, it will amplify Raman scattering, producing a signal that can be detected by the researchers. This way, even single viral particles can be detected, meaning that an illness can be identified in its earliest, symptomless stages. Theoretically, the signal can be amplified by a million times, but this requires special conditions, so in this experiment the researchers only amplified it a thousand times, which proved sufficient.
The resulting spectrum was analyzed with machine learning algorithms, namely the reference vector method that helps differentiate two similar spectra by analyzing those areas that may appear as background noise to the human eye. Thanks to that, it’s possible to not only detect a virus in the sample, but also identify it.
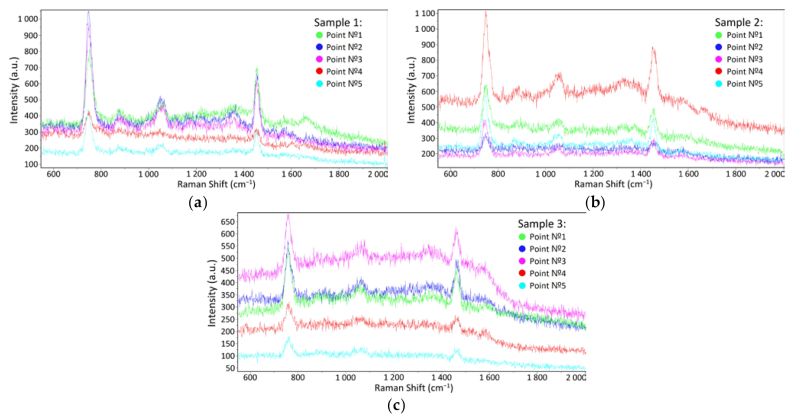
SRES spectra in: a) the pure STE buffer medium; b) influenza A virus in the STE buffer medium; c) influenza B virus in the STE buffer mediumImages courtesy of researchers
The course of the study
Using different viral strains, the experts at the Smorodintsev Research Institute of Influenza grew influenza type A and B viruses and coronavirus in their lab – either in a cell culture or in a chicken embryo.
Once the samples were obtained, they were transferred to a SERS surface with silver nanoparticles, which was then subjected to laser radiation of a 633 nm wavelength. Having scattered in the sample, the laser’s light entered a spectrometer. It took only a minute to detect changes in the laser’s wavelength that signified the presence of a virus in the sample.
Then, spectral data in the form of tables and plots was passed to pre-trained machine learning algorithms designed to detect the presence of a virus and identify its type. The underlying code was edited depending on the algorithm’s performance.
As a result of the experiment, the researchers were able to demonstrate 93% accuracy of the algorithm on the high-concentration samples produced by the institute. With lower concentrations that are closer to those obtained clinically, the accuracy reached 85%. The idea behind the project was to train the algorithm on high-concentration samples to enable it to detect viruses even in lower concentrations.
Production of spectra. Photo by Dmitry Grigoryev / ITMO.NEWS
What’s next
In the future, the team is planning to expand the virus’ database and develop a powerful software for spectral analysis.
“We want to develop a multipurpose system accessible to any research group if they need to detect viruses in any sample. As for more applied use, we would like to develop a device that will help us curb outbursts of viral diseases. For instance, it could come in handy at hospitals or public places: any patient will be able to know if they’re ill within three minutes of submitting a sample,” explain project executive Artem Tabarov, an engineer at the Laboratory for Optoelectronic Support of Cyberphysical Systems, and Vladimir Vitkin, PhD, the head of the project.

Artem Tabarov. Photo by Dmitry Grigoryev / ITMO.NEWS
According to the researchers, the solution can also pave the way for targeted treatment for viral diseases. Counterintuitively, there aren’t many efficient specialized antiviral treatments because to develop those, clinicians would have to identify a specific virus in a patient’s system. This is rarely done for any illness other than coronavirus. In this case, a spectral analyzer can make such tests more accessible, thus producing more data for the pharmaceutical industry about the most common viruses. This data can be used to develop and test specialized targeted treatments.