The research is featured in the Journal of Heredity, a peer-reviewed scientific journal published by Oxford University Press. Pavel Dobrynin, one of the authors and a researcher at ITMO’s Laboratory of Genomic Diversity, discussed what animal genomics is and how it helps preserve and study endangered species.
What are the tasks of animal genomics?
Genomics allows us to gain information both on a specific organism and on the entire population. Basically, the genome is a database – a tool that helps learn more about this or that species.
One of the key tasks of animal genomics (of conservation genomics, to be exact) is to prevent the extinction of endangered species. How? For example, through evaluating risks and demographic history of a population and using genetic diversity as an indicator.
For example, there are few gazelles in wild nature but a lot on ranchos, in zoos, etc. They have preserved a great potential for genetic diversity. So even though there are few of them in nature, we can gradually place them back into the wild and help them adapt, as long as domesticated and wild animals are still similar.

Pavel Dobrynin. Credit: Dmitry Grigoryev, ITMO.NEWS
If there are few animals and inbreeding happens more often, the genetic diversity and the ability to survive decreases. The only way to change it naturally is to help increase the number of animals through breeding. As new generations appear, genetic diversity increases.
Genomes help detect many changes, including potentially harmful ones. If we have an assembled genome, we can try to control some risks both for wild and domesticated animals.
Let’s say, an animal died in the wild due to being susceptible to a pathogen – it’s sad but it won’t harm the population in general. But if the same thing is to happen in a zoo, where animals live together, the disease might spread. It’s genomic information that can lead us to information on a compromised immune system.
How can genomic data be applied in the breeding of domestic animals?
Genomics has greatly benefited agriculture but this field isn’t popular in Russia yet. People started to breed farm animals without knowing anything about genomics and genetics, all they had was some observations on the way animals inherit certain properties from their ancestors.
Over time, domestic animals become more productive: cows produce much more milk than they initially used to. The usage of genomics helps develop selective breeding and thus making it possible to get a specific type of breed in a shorter time. By using genetics and genomic selection, we can say straight away what a cow’s productivity level will be, what diseases it might carry, etc.
Tell us about your work on the assembling of the jaguarundi genome. Is it a type of puma?
Yes, they’re relatives: pumas, cheetahs, and jaguarundis. Jaguarundis look different: they are smaller and have short legs and an elongated body. They mostly hunt small animals in the daytime. While cheetah males don’t especially care about their offsprings, jaguarundis have social structures. There’s not much known about them – they are barely studied in Russia.
Cheetah and puma’s genomes are already assembled. Jaguarundis are their closest relatives. When it comes to preservation, however, they aren’t endangered.

Puma and cheetah. Credit: depositphotos.com
This species was interesting for us because they’re similar to other cats but have their own distinctive features, too. These three types of cats hunt, live, eat, and even look different to each other.
By assembling the genome, we realize what characteristics an animal has, why it behaves in a certain way, why it looks the way it does, and what’s the molecular and genetic foundation behind all this. Our article will help further research of jaguarundis, as other researchers will be able to use our results.
Where did you find the jaguarundi genome?
Jaguarundi isn’t a rare or protected species so it’s easier to find and deliver its genome. Our colleagues from the Institute of Molecular and Cellular Biology helped us – they had samples of frozen tissues. Then, one of the samples was sequenced. This sample went a long way: from Novosibirsk, it went to Johns Hopkins University in Baltimore, where the data was obtained. Then it was sent to us and we assembled it.
How did you do that?
Once the genome is sequenced, we end up with hundreds of millions of small fragments that need to be assembled like a jigsaw. We collect various genome elements, mark them, and evaluate the quality methods and genetic diversity of the result. Genomes of jaguarundi haven’t been assembled before – we did it first.
We plan to conduct a comparative analysis of jaguarundi, cheetah, and puma. The idea is to learn more about the biology of these species by using high quality genome samples.

Jaguarundi. Credit: Gelia Sudzilovskaya, ITMO’s Laboratory of Genomic Diversity
What did you learn from the sample?
We have thoroughly studied the structure and organization of the jaguarundi genome; there aren’t many cat and mammal genome assemblies with such precision. Just as we expected, the organization of the genome is very conservative among all cat species. Our further work lies in a thorough study and description of differences between various cats. First things first, we’ll compare jaguarundi with African cheetah and puma.
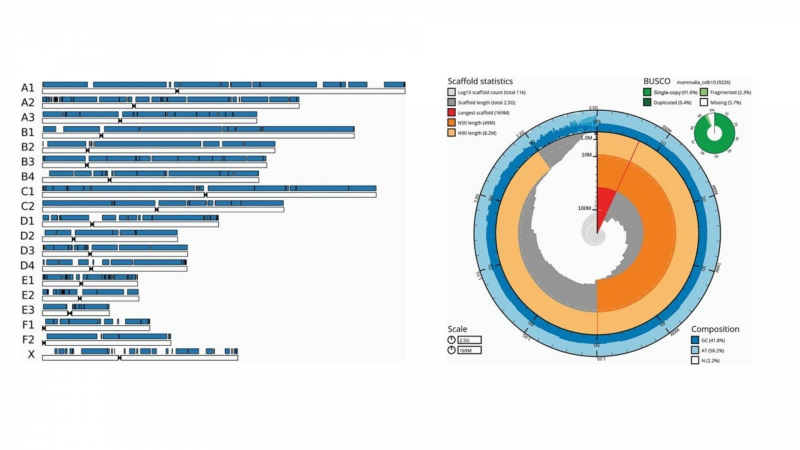
On the left: Scaffold fragments from the jaguarundi's genome assembly placed on domestic cats' chromosomes. On the right: jaguarundi genome assembling. Credit: the article from the Journal of Heredity / https://academic.oup.com/journals
The genome looks quite healthy and genetically diverse; we haven’t detected any potentially harmful mutations. Once the genome is published, it becomes available for researchers worldwide and perhaps someone will find out more while we’re busy with the comparative analysis.
So genomes are open-access materials for researchers?
Yes, exactly. Many assembled genome sequences are in public access. Each new genome increases our opportunities for analysis of the species and other genomes. The more data we have, the better are our results in the fields of comparative analysis, phylogenetics, and evolution.
By having a genome, we can create marker panels for research of population structure, demographic backlog, and other work in the field of population genetics. Sometimes genomic data helps create tools for the control of illegal hunting. I’m sure we’ll see more and more creative applications of this information.