A conscious approach to treatment
As estimated by the World Cancer Research Fund International, more than 18 million people worldwide were diagnosed with cancer in 2020, with skin cancer (melanoma) being the 17th most common type of cancer.
Nowadays, cancer treatment types include surgery, chemotherapy, radiotherapy, hormone therapy, as well as immunotherapy. While the first four methods are meant to destroy cancer cells, immunotherapy, on the other hand, boosts a person’s immune system to fight the disease. It uses immune checkpoint inhibitors to rev up immunity so that T-cells could destroy cancer cells that the body left with no response.
Although immunotherapy is considered one of the most promising therapeutic approaches against melanomas at the inoperable or metastatic stage, the data concerning the treatment’s effectiveness is often contradictory: while some find the method effective, some experience no response, others have to deal with a number of severe autoimmune side effects.
Moreover, medical specialists have no tools that would allow them to predict the efficacy of the treatment and understand which factors affect its outcomes. As noted by Artem Ivanov, an employee at ITMO’s International Laboratory "Computer Technologies", one of the possible solutions can be found in intestinal microbiota. The fact is that bacteria that reside in the human intestine are important to the function of the immune system: for instance, they help produce antibodies and lymphocytes meant to neutralize viruses and pathogens.
“In case of the immunotherapy failure, patients can receive a fecal transplant that will change the composition of their gut microbiota and induce their immunity. Yet, the biggest problem with transplantation is that there are no criteria to follow when selecting a donor for each individual patient. Hence, we want to come up with a method that would, for instance, help specialists find donors with certain microbiota or manually produce probiotics containing beneficial bacteria,” explains Artem Ivanov.
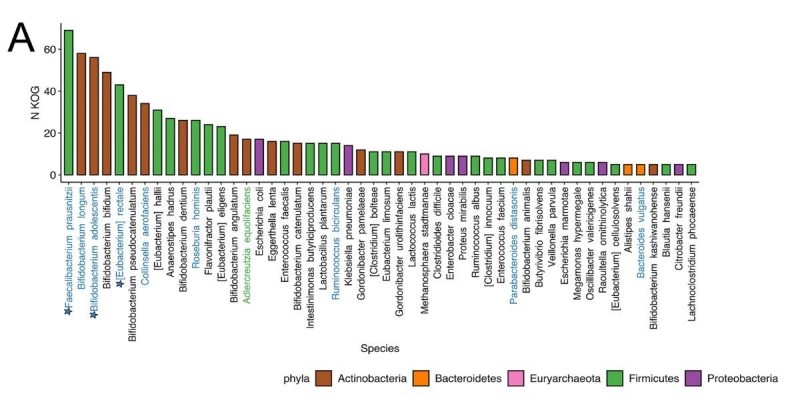
Species rankings among patients with positive and negative responses to immunotherapy. Species associated with several studies are indicated with stars and colors. Illustration from the article / journals.asm.org
Looking for a bacterium in the microbiota
To make it happen, the researchers from ITMO’s International Laboratory "Computer Technologies" and their colleagues from the Lopukhin Federal Research and Clinical Center of Physical-Chemical Medicine conducted a large-scale study with a view to identifying biomarkers and gut bacteria that can indicate whether the treatment will be successful or not.
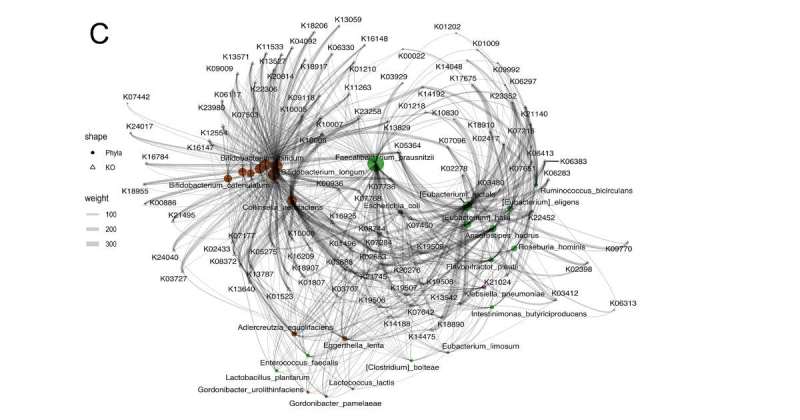
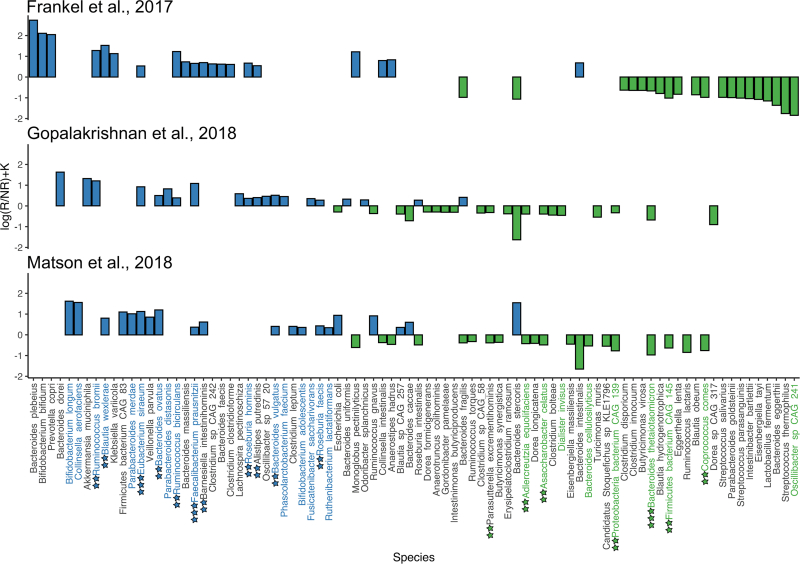
The study involved 680 gut metagenomic samples from patients with oncological diseases and varying responses to immunotherapy. All the data was taken from seven studies published in an open database of the National Center for Biotechnological Information. Having thoroughly investigated the cases, the researchers divided them into three groups (metagenomic samples from immunotherapy patients and those who received or were going to receive a fecal transplant) and then compared the information. For that, they employed their own algorithms MetaCherchant and RECAST, as well as the Songbird algorithm. MetaCherchant defines relationships between bacteria and genes of interest, RECAST studies metagenomic samples from patients before and after the transplantation, and Songbird is used to analyze compositional data.
As a result, the scientists managed to identify 101 donor-derived functional biomarkers that can determine a patient’s response to immune checkpoint inhibitors (ICIs) therapy. The data obtained can be used to develop a diagnostic test that can predict therapy success before starting the treatment.
Apart from the immune-related biomarkers, the researchers found three types of bacteria – Faecalibacterium prausnitzii, Bifidobacterium adolescentis, and Eubacterium rectale – in the composition of the patients’ intestinal microbiota. This data can be further used to build a personalized program to improve gut microbiome before immunotherapy, for instance, if a patient lacks certain gut bacteria vital for treatment success.
“It is very rare to see the same bacterium in different studies. Sadly, one bacterium can't tell whether a person is sick or not. That’d be too easy! The human intestine is much more complicated, so most of the time, we have to look for combinations of bacteria and biomarkers. While the presence of a certain bacterium may not mean anything, the fact that it releases substances that feed another – does. In this sense, what we do is try to make our search for a single bacterium in a haystack-like microbiota more efficient with the use of the algorithm,” says Artem Ivanov.
See also:
Analyzing Metagenome Helps Understand the Role of Bacterial Species in Crohn’s Disease
Efficient algorithms for gut analysis
In order to validate the findings of the study, the specialists from the Lopukhin Federal Research and Clinical Center of Physical-Chemical Medicine intend to run several tests, including on mice. Within the experiment, they will transplant melanoma cells to mice and use the bacteria theory to treat them. If successful, the scientists will be able to develop specialized digestion fibers and probiotics to improve a patient’s gut health and thus enhance the effect of immunotherapy.
In the long run, the developers are planning to make their database open for other researchers seeking to find and analyze metagenomic biomarkers that are (in)efficient for immunotherapy. According to Artem Ivanov, the methods used in the research are also applicable for other types of malignancies, including non-small cell lung cancer or cervical cancer. Yet, the key mission of the researchers is to develop computational methods for a wide range of gut microbiota-related studies.
“There are several hard-to-diagnose inflammatory bowel diseases, such as Crohn's disease and ulcerative colitis. As of now, we are running experiments on open-source data to potentially use it to analyze microbiota and thus help medical specialists improve diagnostics and prescribe more accurate treatments, if needed. In addition to that, we also consider other fields, for instance, microbiota and neurodegenerative diseases, in particular Alzheimer's disease, were also found to be connected,” notes Artem Ivanov.
The study is supported by the Russian Science Foundation and Priority 2030 Program.
Reference: Evgenii Olekhnovich, Artem Ivanov, Anna Babkina, Arseniy Sokolov, Vladimir Ulyantsev, Dmitry Fedorov, Elena Ilina. Consistent Stool Metagenomic Biomarkers Associated with the Response To Melanoma Immunotherapy (mSystems, 2023).