Over the course of our lives, we are constantly surrounded by bacteria – apart from those of our own, we encounter them in the air, soil, water and even in our homes. Some of these external microorganisms can be harmful to humans, while others play a vital role in regulating our immune system.
With urbanization taking over the world, the situation is changing: people are still surrounded by bacteria, but their diversity is now decreased and we are exposed to them to a lower extent. On the one hand, cities have smooth surfaces, which are regularly cleaned and sanitized. On the other hand, there is public transport, there are schools, offices and factories, libraries and gyms which serve as a platform for active bacterial exchange between many different people. Several years ago, an international group of researchers set out to explore the microbiome of subways, the vital transportation systems of modern cities. New York was first to have its subway microbiome investigated.
“The researchers analyzed the floors, handrails and carriages, and found a curiously varying microbiome,” explains Alexander Tyakht, a PhD in biology, the chief technology officer at Knomics (Atlas R&D), and staff member of ITMO University’s Computer Technologies Lab. “An international consortium was later established to study the subway microbiomes all over the world. This project aims to catalogize the diversity and patterns in their microbial ecology, thus creating a base for a more focused analysis of microorganisms inhabiting public spaces.”

It’s all in the method
Traditionally, to study the microbiome of a certain surface you would need to cultivate samples from it. To put it simply, you have to take a swab sample from the floor, the wall or the handrail and streak a Petri dish with the swab. There, the microbes reproduce, which makes them easier to examine.
This method, however, has a significant drawback: without prior knowledge of what kind of bacteria were collected it is difficult to choose a suitable medium for them to flourish – some microorganisms need light while others don’t, some can’t tolerate air and some suffer in salinity. Moreover, many microorganisms are practically uncultivable. As a result, some of the collected bacteria will not reproduce in a dish, and the researchers will have no way of establishing their presence on the sampled surface.
“However, if we use high-throughput DNA sequencing, it allows us to detect a greater diversity of bacteria from the sample, even if we can’t culitvate them in the lab,” continues Alexander. “Certainly, the method is not without limitations. For instance, it does not tell the absolute number of bacteria on the surface neither which of them are viable. But we can still evaluate their total diversity and identify exactly, sometimes down to the level of species, what kind of bacteria are included in the community. A similar method was used in New York subway study.”

The inhabitants of the Moscow Subway
The same method was used by Russian researchers to look into the microbiome of the Moscow Subway. Alexander Tyakht, together with Natalya Klimenko, a bioinformatician at Knomics (Atlas R&D), and Dmitry Alexeev, a staff member of ITMO University’s Computer Technologies Lab, have analyzed samples from handrails, floors and other surfaces using microbiome sequencing. The data was analyzed with the Knomiks-biota system and is available online.
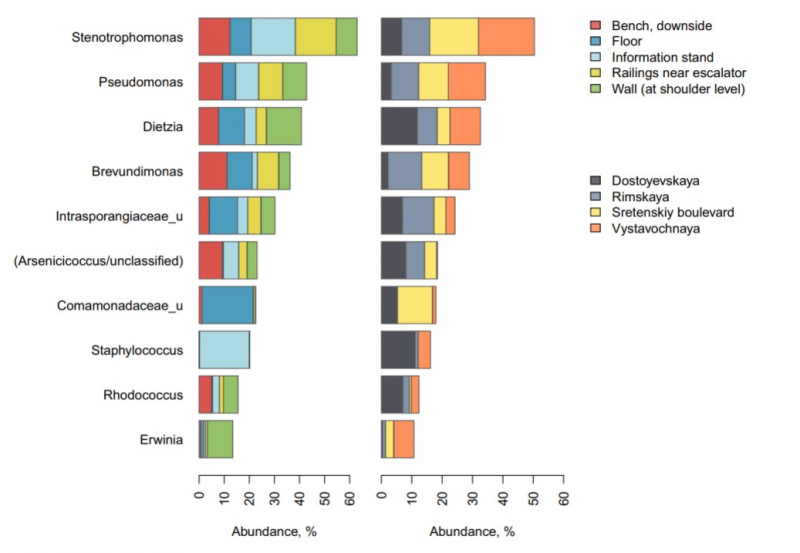
“The majority of microorganisms that we found fall into two distinct categories – it’s either human skin bacteria (previously observed on the skin) or soil bacteria (found in soil and on plants),” explains Natalya Klimenko. “Most surfaces had a mixture of these two groups. Only the information stands had greater proportions of skin bacteria. We observed that there were stable combinations of skin and soil bacteria which co-occur. Possibly, there are certain emerging ecological interactions between these bacteria that may be be relevant to human health. Of course, we cannot claim there is a certain symbiosis based on our project alone, but now we have prioiritized a list of bacteria to be analyzed with more advanced methods.”
After comparing their results with those obtained by the US researchers, the Russian group discovered that the most common types of bacteria in the subway systems of Moscow and New York are similar.
“Most of the Dietzia, Brevundimonas, Pseudomonas, Arsenicicoccus, Stenotrophomonas and Brevundimonas - highly abundant in the Moscow Subway - have also been detected during the previous New York project,” says Natalya Klimenko, a bioinformatician at Knomics (Atlas R&D). “Apparently, the subway microbial community structures manifest similar patterns across the globe.”

One other interesting outcome was the identified link between the microbiome diversity and the passenger traffic across the stations. At the same time, the highest diversity was found in places with abundant soil bacteria, while the prevalence of the skin microbiome corresponded to lower diversity. This pattern resonates with the fact that in nature soil microbiome is richer than that of our bodies.
Noteworthy, the researchers found no significant evidence of pathogenic bacteria: no DNA sequences were reliably detected for any of the ten pathogens that could be robustly detected given the applied method. Nevertheless, the researchers emphasize that this finding doesn’t mean you can now stop washing your hands after a subway journey: only a handful of samples have been analyzed, and the analytical method cannot detect all known pathogenic bacteria, viruses, fungi or protozoa.
Bacterial residue
According to the researchers, the current study of the Moscow Subway is only a pilot project meant to demonstrate how DNA sequencing can be used to study the microbiome of public places. Hundreds of samples from every station and every possible surface are to be analyzed for a more global mapping of the Subway’s biological diversity. While such a project is too expensive now, it can be brought to life in the future.
“Such dense sampling will allow to create a detailed spatiotemporal map of the public transportation system’s microbiome,” says Alexander Tyakht. “In the prespective, by analyzing these Big Data, it will be possible to mine the information about the human flow around the city and the patterns of their interaction with the environment. This map would be even more exact if we could observe the time dynamics of microbiome composition over time. For the current project, we collected all our samples in one day, but there might be bacterial waves changing with the time of day, such as the rush hour. Finally, there is the famous Ploshchad Revolyutsii station with its hand-polished statues. Everyone rubs the nose of the patrolman’s dog there for good luck. It would be interesting to see how the statue’s microbiome changes over time – daily, in winter, spring or when the students are about to have their exams.”

Reference: Natalia S.Klimenko, Alexander V.Tyakht, Stepan V.Toshchakov, Margarita A.Shevchenko, Aleksei A.Korzhenkov, Ebrahim Afshinnekoo, Christopher E.Mason, Dmitry G.Alexeev, Co-occurrence patterns of bacteria within microbiome of Moscow subway, Computational and Structural Biotechnology Journal, Volume 18, 2020.