Starting from 2016, the expert board of the Systems Biology Program annually selects the five most promising young scientists under the age of 35 years engaged in systems, molecular, and cell biology, bioinformatics algorithms and methods, comparative bioinformatics, genomic research, and so on. The mission of the program is to support and stimulate Russian research in related fields.
Ekaterina Noskova became the third ITMO scientist – following the footsteps of Alexey Sergushichev and Konstantin Zaytsev – whose work was highly regarded by the expert council of the scholarship program.
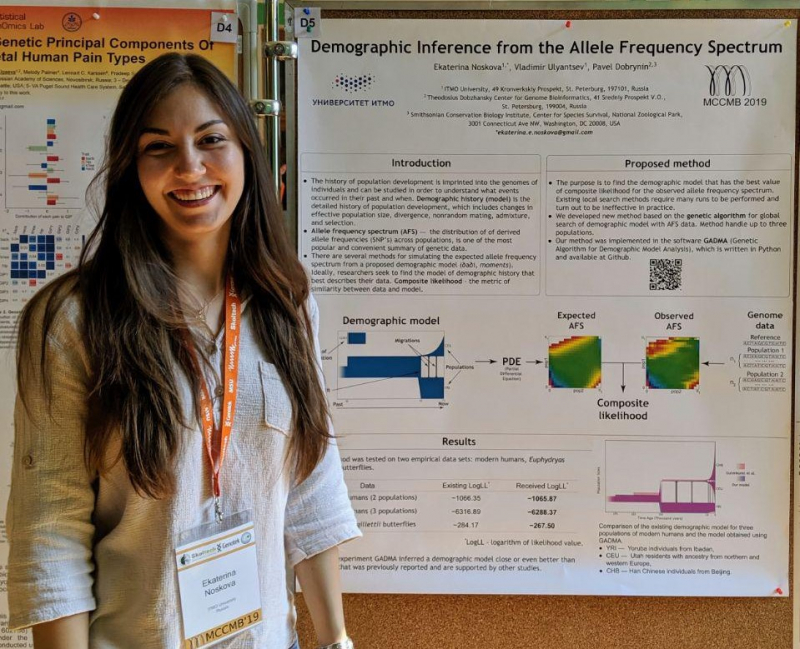
Demographic history
The researcher received a 600,000 rubles grant (with annual prolongation for up to three years) to develop a program for inferring the demographic history of populations, both human and animal. The GADMA (Genetic Algorithm for Demographic Model Analysis) is able to reconstruct the evolutionary history of three populations simultaneously and can be used to conduct research using methods based on the allele frequency spectrum (AFS).
“The demographic history of any population includes data on the evolution of the species: how populations changed over time, where they migrated, and whether they were exposed to natural selection or not. Researchers can use genomic data to reconstruct the story. This way, they can track, for instance, a gap between the populations of different continents or, when we talk about humans, to find out that they left Africa 150,000 years ago,” explains Ekaterina Noskova.
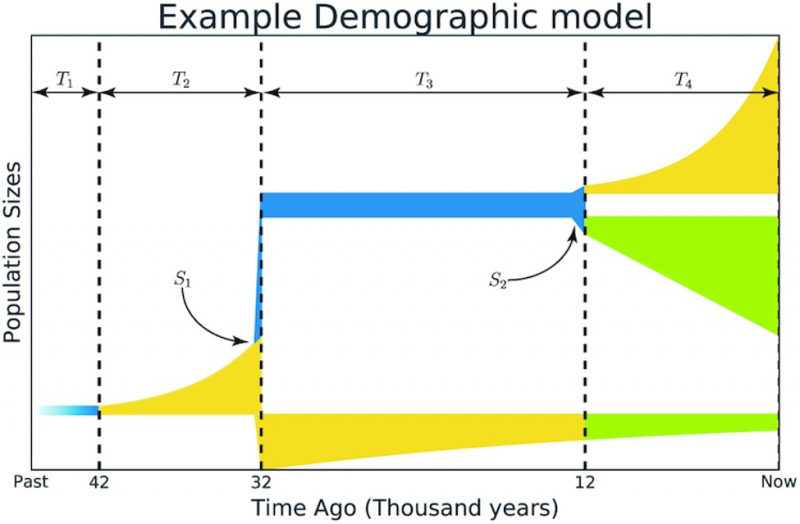
Illustration from the article in GigaScience. Credit: academic.oup.com
At the moment, there are two commonly used simulators for building demographic histories from AFS data: dadi and moments. Both, however, have certain limitations. Based on a mathematical model, they don’t perform well with overlapping generations and generally give less accurate results. The GADMA program, in turn, allows scientists to switch between two simulators and get the best of both choices.
“Dadi is one of the most popular methods based on what is called the allele frequency spectrum. It is genetic data presented in a simplified manner – matrices. After all, it is hard to work with complete genomes as they contain too much information,” shares Ekaterina Noskova.
The developed program provides a choice of two simulators and automatically selects a suitable model for working with certain data. Basically, it cycles through all the possible combinations, compares simulations with given results, and concludes their probability,” comments Ekaterina Noskova.
How does it work?
Let’s say, researchers collected genetic data on cheetahs, which they can now transform into a rather simple model using AFS-methods or other statistics, for instance, haplotypes. Then, they have to choose. They can take one of the ready-made solutions like dadi or moments. If so, they will have to figure out how each program works and, while doing it, they will inevitably learn that their optimization algorithms may fall short of their expectations.
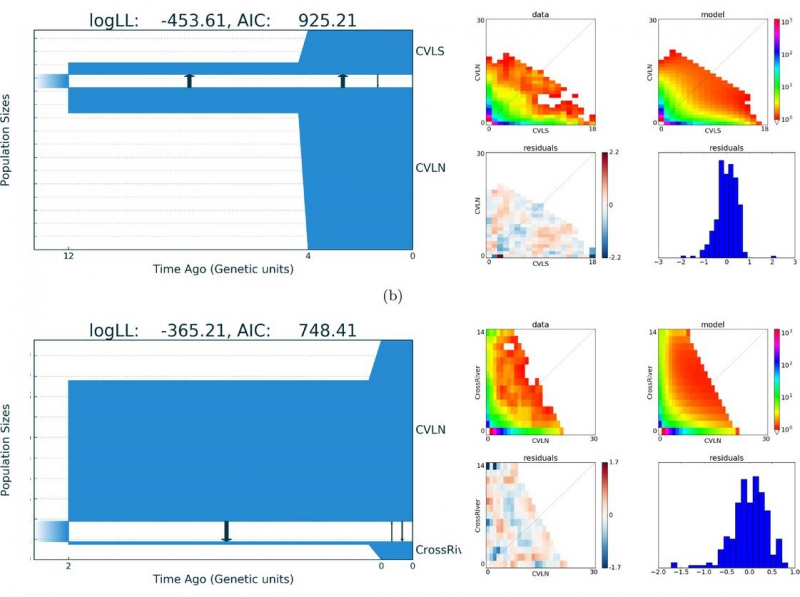
Illustration from the article in GigaScience. Credit: academic.oup.com
GADMA, in turn, offers a semi-automatic mode. Users don’t have to thoroughly study each simulator, they can simply choose a preferable one, upload their data in the program, and receive a finished demographic history, as well as its visualization and code. This algorithm is more accurate and shows more stable and realistic results.
The creators plan to completely automate the selection process, expand the program’s functionality, and add a new AFS simulator – fastsimcoal2 or momi2. Future plans also include the integration of simulators based on haplotypes, for example, diCal2. Although such simulators can handle large datasets, they are much slower. The scientists believe that the combination of faster and more accurate methods could yield very useful results.
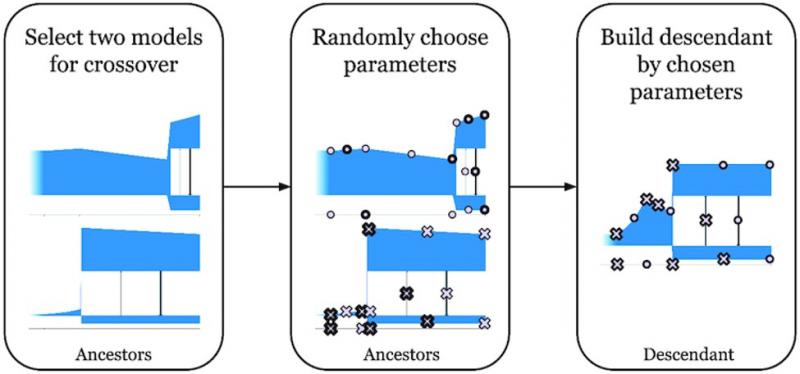
Illustration from the article in GigaScience. Credit: academic.oup.com
The history of creation and future plans
As Ekaterina recounts, she started the GADMA project in 2017 when she studied algorithmic bioinformatics at St. Petersburg Academic University. She came up with the algorithm and prototype during the BioHack hackathon. The task was to infer the demographic history of cheetahs using the dadi simulator.
“We noticed that the simulator didn’t handle the situation very well, so I tried to come up with a new algorithm just to see if it’d work better. And it did. The algorithm turned out to be quite efficient, and I continued to work on this project, wrote a Master’s thesis, and then did my internship at ITMO’s International Laboratory for Computer Technologies. Together with Vladimir Ulyantsev and Pavel Dobrynin, we've been developing this program for several years now. And, in early 2020, we published an article in GigaScience,” says Ekaterina Noskova.

Ekaterina Noskova and her team at BioHack 2017
The researcher applied for the grant last year, too. Learning from the past mistakes, she expanded her portfolio, conducted additional research, and drew up a more thorough development plan for the next three years.
The Systems Biology Program runs for three years, with a total grant being 1.8 million rubles. Even though grant holders don’t need to regularly report on their spending, they deliver an annual presentation at Skoltech’s seminars held specifically for the program fellows. The final report, with the results of the three-year work, will take place at the Philip Morris International research center in Neuchatel, Switzerland. This year’s winners will also present their projects at the Moscow Conference on Computational Molecular Biology (MCCMB), which will be held from July 30 to August 3 at Skolkovo Innovation Center.