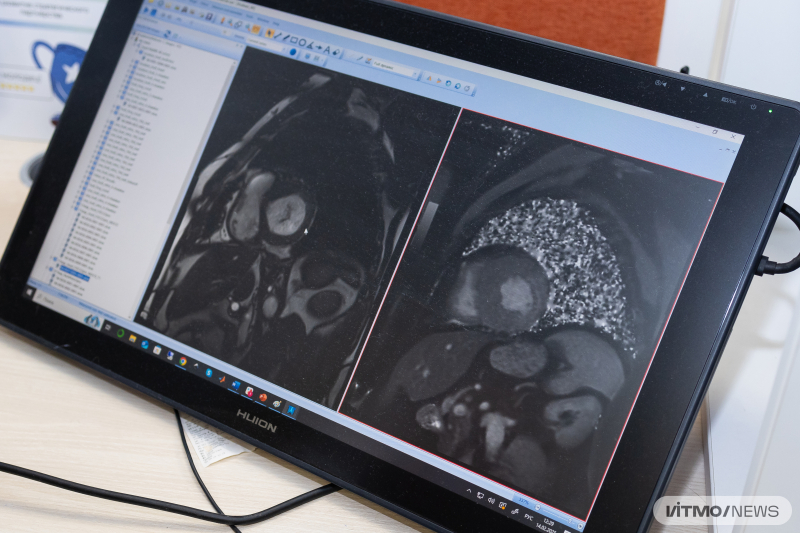
Cardiovascular diseases are the leading cause of death globally. For timely detection, it’s important to conduct diagnostics, including locating fibrosis – scar tissue that can appear after heart attacks (postinfarction cardiosclerosis) or infectious diseases. One promising screening method is MRI, which is minimally invasive and uses non-ionizing radiation.
In diagnostics, morphometry, the precise measurement of fibrosis volume, takes a lot of time – it has clinicians manually determining the approximate percentage of fibrosis tissue in a heart segment; the percentages are used to fill in a table that, in its turn, is used to create a 17-segment diagram. On average, processing one image series takes between one and two hours per patient.
Neural networks can help cut down on the processing time, however the existing models aren’t accurate enough and require manual or semiautomatic selection of fibrosis areas. This has researchers developing methods for automated generation of the 17-segment diagrams based on MR images.
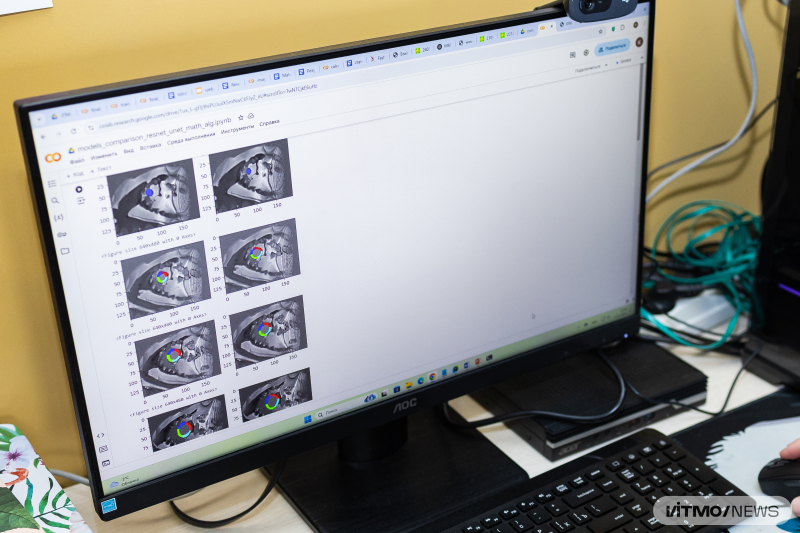
For this purpose, researchers from ITMO and Almazov National Medical Research Centre have developed a semiautomatic deep learning model that tackles the problem in stages: first, it locates the cardiac muscle; then detects the fibrosis within each of the 17 traditional heart segments and evaluates its amount in each of them.
“For the suggested algorithm to work, users only need to mark several points on a cardiac image and classify the cross-sections; the tissue segmentation and 17-segment diagram generation are fully automated. We are now working on improving our method, aiming to develop a faster, fully automated algorithm that will be able to analyze images instantaneously and without user assistance,” shares Walid Al-Haidri, the main executor of the project and a researcher at ITMO.

Ekaterina Brui, the head of the project and senior lecturer at ITMO's Faculty of Physics, and Kseniia Belousova, the project's executor, an engineer at ITMO's Faculty of Physics. Photo by Dmitry Grigoryev / ITMO.NEWS
While it takes a clinician about one to two hours to process an image, the model can do it in a few minutes. Moreover, it can work with an image in a single plane, while clinicians may require several images in different planes for their diagnostics – which means a longer MRI scanning procedure and its subsequent analysis.
The researchers used the U-Net neural network and a cascade algorithm. The model was trained on manually marked cardiac images, as well as on post-heart attack MR images from University Hospital of Dijon (France). Overall, images from 250 patients were used in training.
Currently, the algorithm’s output matches the opinion of two experts in 86% and 77% of cases respectively, which the authors see as a good result – typically, experts agree about 80% of the time, which means that the model has human-level performance.
Based on the data on fibrosis amount and localization, clinicians will be able to quickly and accurately predict heart function complications and disease outcomes, track the dynamics of the heart’s condition, and come up with more efficient treatment strategies. In the future, the algorithm could be used not only to process MR, but also CT images.
“We don’t just use big data to train neural networks to perform routine tasks; instead, we are offering a tool that can help solve complex tasks at the level of an experienced clinician and acquire more information on the connection between fibrosis localization and other parameters of the heart,” says Ekaterina Brui, the head of the project and a senior researcher at ITMO’s Faculty of Physics.
This study is supported by the Russian Science Foundation grant No. 23-75-10045.