How to determine when modern tigers first appeared on Earth? When did the two elephant populations split? Is there a difference between the Dama and the Moroccan gazelle? When did the division of the African and the Eurasian Homo Sapiens occur? The answers to all these questions can be found in the population’s demographic history: a scenario that shows what stages a population went through in the course of its history, whether it underwent any mass extinctions, migrations, or sharp spikes in its numbers.
“Before people learned to decode genomes, the only way to find out what happened to populations of people, cats, dogs, elephants etc. was paleontology,” explains Vladimir Ulyantsev, associate professor at ITMO University’s Information Technologies and Programming Faculty and head of the Computer Technologies international research center. “We could see how many fossils of the studied species there are in each layer of the soil. But this opportunity is limited, the finds are limited, and the assumptions made on their basis aren’t always accurate. Then, people came up with genome sequencing methods, thus obtaining genetic data in addition to the existing one. For example, any organism’s genome keeps traces of the events that happened to its ancestors in the past. Migration, decrease or, vice versa, increase of population size – all this leaves some kind of a mark on DNA. Using a lot of different statistics and other methods, based on changes in the frequency of the alleles we can reconstruct the processes that transform the genome into what we can see now.”

This information can help us find out when the groups of our ancestors divided into the indigenous populations of, for example, the Pskov and Ryazan regions. Or when mustangs became feral again. But in addition to fundamental questions of population genetics, this data can contribute to the research in the field of ecology and environmental protection. For instance, if some region has only 800 walruses left, scientists have to understand whether it constitutes a critical decrease or it is a natural size of the population, which has remained constant for several thousand years, and answer the question of necessity in usage of valuable resources to protect this species from extinctinction. This requires re-tracing the population’s demographic history.
Genetics and informatics
Modeling of a population’s demographic history from the genetic information is a complicated task which requires population geneticists to know biology and also have programming skills. Scientists have to gather data and write a code for computing possible models of a population’s evolution, which could have led to the genetic information we observe in this population’s individuals today.

“How was the demographic analysis conducted up until recently?” explains Ekaterina Noskova, member of ITMO University’s Computer Technologies international research center. “Scientists used different programming software tools that allow them to measure the probability that demographic history with its parameters have led to the observed genetic data. Thus, the scientists searched for a demographic history that corresponds the most with the real-life data. The programming software provided some optimization algorithms for searching for the parameters of the predefined demographic model, but these were local optimizations, which means that they required some initial parameter values, very strongly depended on the initial solution and only improved it in a neighborhood of initial point. There are also restrictions imposed by the researcher’s choice of the model itself.”
Therefore, the process of demographic history inference was long and required specialized knowledge and skills both in programming and genetics. But most importantly, the final result very strongly relied very heavily on the researcher’s initial hypothesis.
“The scientist’s work consisted in coming up with various scenarios, choosing the most probable ones and using the optimization methods. Then the program showed which of these scenarios is the most likely. And the existing optimization algorithms couldn’t find any scenarios beyond those they were offered,” says Pavel Dobrynin, member of the Computer Technologies international research center.

Optimization of solutions
The software developed by a group of ITMO University scientists as part of the Project 5-100 grant programs and with support from JetBrains Research aims to solve this problem. The researchers proposed a programming product that independently and automatically predicts the most probable demographic history of populations. It is also significantly less dependent on the initial research hypothesis, doesn’t require advanced programming skills and produces more accurate results. It is also quite flexible, which means that if the obtained result somehow diverges from archaeological or historical data, you can easily introduce additional limitations into algorithm, and it will update its hypothesis.
“Using genetic data, our software automatically finds the model it considers optimal,” shares Vladimir Ulyantsev. “It looks at the entire space of all available scenarios. As a scientist, I’ll sort the scenarios and choose the one I deem most likely: There may be three, five, maybe ten of them. The software, on the other hand, will test all of the models it considers probable, which is a much larger amount. That’s why its solutions are better than those proposed by people using the usual methods. The most beautiful thing here is the method – a genetic algorithm inspired by evolution: individuals multiply, mutate, the least adapted die out. In our case individuals are demographic models with parameters, and their adaptation is determined by the similarity to the observed data.”
After obtaining this data, the scientists can map it and compare the information about when that population underwent a migration with archaeological findings and other evidence.
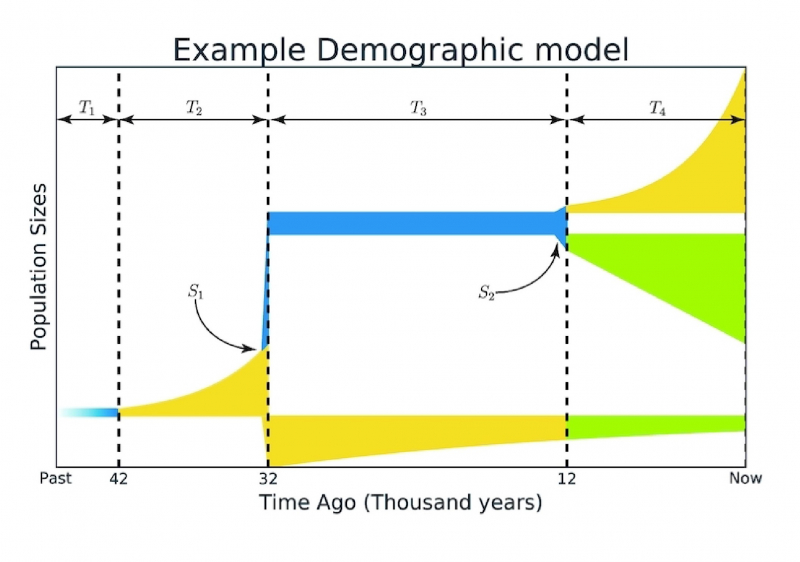
The proposed method was used to check a large number of existing hypotheses and research by evolutionary geneticists. In many cases, the obtained result was much more accurate than in the original work.
“When we were inferring demographic histories for various populations, our algorithm worked better than the original research,” points out Ekaterina Noskova. “It was able to predict a model that provided better measure of similarity with the same data. For example, we analyzed one paper where 83% of models turned out to be under-optimized. Also we looked at the data of modern humans in order to get more information about the division of Homo Sapiens into the African and Eurasian populations. The model we obtained as a result had a much better conformity to the data than previously. Our findings demonstrated that a population of humans left Africa around 150 thousand years ago, and the size of Eurasian population wasn’t constantas it was previously thought. Instead, there was exponential growth.”
The algorithm proposed by the scientists has already been experimentally tested in a range of research initiatives, including those conducted as part of the Russian Genome project. Specifically, with its help scientists managed to establish the common history of the inhabitants of Pskov, Novgorod and Yakutia.
Reference: Noskova E, Ulyantsev V., Koepfli K-P., O’Brien S.J., Dobrynin P. Genetic Algorithm for Automatic Inferring the Joint Demographic History of Multiple Populations from Allele Frequency Spectrum. GigaScience, 2020 / 10.1093/gigascience/giaa005





