On studying at ITMO University
Why did you decide to join ITMO specifically?
When I finished school, I had a poor understanding of whom I wanted to become, I just knew that I was interested in maths. I was a competition winner in this subject, so I could go anywhere. I got interested because there was a Department of Advanced Mathematics at ITMO, where they had the Applied Mathematics and Computer Science specialization. Several months before admission, I went to ITMO for an Open Day. I liked the people who organized everything at the department. After talking to them, I thought that studying there would be interesting.
What were your impressions when the studies began?
The first two years of the Bachelor’s studies were very interesting. There really was a lot of advanced mathematics at the Department of Advanced Mathematics and it was great. However, we also had programming courses, which got me interested in programming and in the end, I became more interested in that than in math.
At ITMO, everything is organized in such a manner that after the second year, students get more freedom. They are encouraged to work for third-party organizations, so in my third year, I started working, much as most of my peers. By the end of my fourth year, I understood that I still didn’t know everything I wanted in my life, and started to consider doing a Master’s program.

I had to make a choice: either to join a regular Master’s program that I could combine with my work, or join a hard and time-demanding program. I chose the latter. That was in 2018, when in the matematician and programming community of St. Petersburg, everyone knew that the best Master’ program on applied mathematics and programming was at the Academic University.
And right at that time, it moved to ITMO - now it’s a corporate program with JetBrains. I joined it, and it turned out to be very difficult. ithout doubt, this was an extremely interesting and captivating experience. But the most important thing is that the knowledge I got are really relevant, I make use of them on a daily basis.
What is it that you remember most about this Master’s program?
The best thing about it is that you don’t just listen to a course and then forget it. You become confident that you really know something and can apply this knowledge. It often happens that a course includes complex things and fascinating topics, but you forget everything after you pass the exam. In our program, you can forget some things after the course is done, but you definitely know that you possess the necessary instruments, you know where to find relevant information and you know how to apply it all. In short, you are confident in the knowledge you got, and that’s cool.

How to get a job in your field during your third year
You said that you got a job when you were still a student. Was it some part-time job that had nothing to do with your field, or you got the chance to apply your knowledge?
My first job was at BIOCAD, where I still work. During my second year, I had a lecturer, Pavel Andreevich Yakovlev, who was then the head of the Computational Biology Department and is now responsible for everything related to early stages of development at BIOCAD. He invited several students, me included, for an internship. We were given tasks that had more to do with structural biology, but the approaches to solving them were that of mathematics and analysis. We had to analyze data and do modeling. In my third year, I did another internship at BIOCAD, which had more to do with programming: the company needed another analytical tool. After that, they gave me a position.
But you had to take a break from your job in order to do a Master’s program?
Not really. I have good relationships with the management, they are my lecturers at ITMO. So I just had to go part-time instead of working full-time. My studies were a priority, but I worked two days a week. They are ok with it, I’m not the only one here who’s doing something like that.

So what about your career path during this time?
First, I was a data analysis specialist. In my second year, I had a course on that, where they gave us a good amount of knowledge which helped get a good perspective of this field. That’s what made it possible for me to come to work and apply what I learned to real tasks. Then they started to involve me in algorithmic projects and structural bioinformatics, and this is how I gradually became a developer of bioinformatics software.
Structural bioinformatics is basically a treasure trove of interesting computational tasks that are often solved using algorithmic approaches.
How algorithms help in drug development

What are you currently doing for your company?
To put it simply, there are two ways to develop a drug. The first option is to just make a team of scientists synthesize lots of molecula and choose the ones that have a therapeutic effect. The other is called rational drug design. You do computer modeling, and its results allow you to focus on fewer molecules that are known to have some beneficial properties.
Modeling of biological processes is a complex issue, it involves lots of various tasks, and almost all of them have to do with algorithms. For example, one of the stages has to do with predicting the structure of a complex that would be formed by the drug molecule and the target molecule with which our drug will interact in the human body. Studying how this complex will look is called the docking task. I am currently developing an algorithm for this very purpose.
Let’s say there’s a malignant cell in a human organism. We can make a molecule that will interact with it and by doing that, attract the attention of the immune system and make it understand that this is a bad cell that has to be destroyed.
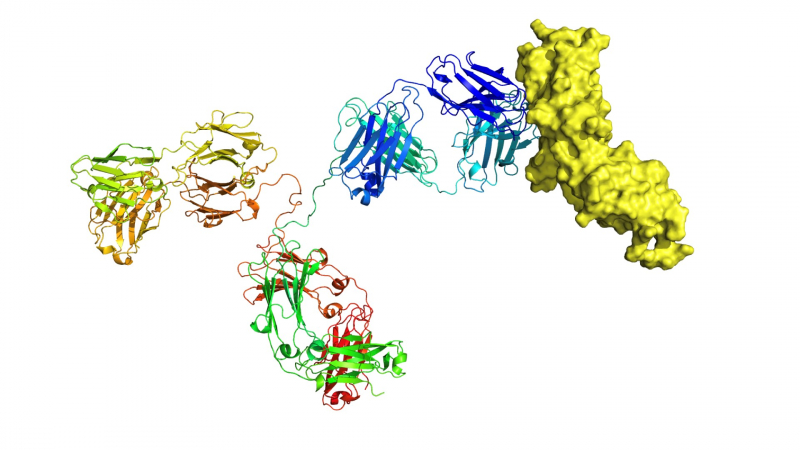
Another example: we know that one of the types of hemophilia is induced by the absence of a specific protein in a human body, which helps interact with other proteins: as a result, blood doesn’t clot. In very simple terms, we can create a replacement for this protein that would bind the necessary proteins and therefore cure people from hemophilia.
In essence, these two examples are about molecular interactions and their structure. This structure can be predicted using computational means. I develop an algorithm for this purpose.
And how does it work?
In the initial approximation, everything comes down to a common enumeration: we have one molecule, we have another, and we want to find the position of the first in relation to the second. So we exhaust all possible options. Nevertheless, this enumeration is not so simple: we apply various heuristics in order to decrease the search space.
The point here is that we can’t perform the enumeration as it is: it takes too much time, and when it comes to drug development, we may need to repeat this process a hundred times. That’s why it’s important to work fast. There are special methods that make it possible to decrease the search space. The docking mechanism operates with the help of these methods: it consists of three consecutive stages, each with its own heuristic that allows to decrease the search space in such a way that no correct answers would be lost.

You took a Master’s program with JetBrains, so you can give a comparison. What’s the difference between being a programmer at a pharmaceutical company and in IT?
If it’s algorithmic bioinformatics that we’re talking about, the field that I work in, here we have a lot more research work. If you work for a major IT company, you have some templates that you have to work in accordance with. If you write some plugin, then chances are that there are millions of similar ones, you just need to code it. Sure enough, sometimes you can be creative: you always have to optimize one thing or another, but you usually work with tasks that are well-known.
In research projects, such as the ones in algorithmic bioinformatics, you often don’t know what to do and how to do it. You start everything with a literature analysis that can take a month. Within this time, you need to learn how the task is being solved for the time being, and whether you need to come with an alternative solution or upgrade the existing one. Then you create an algorithm. In the process of doing all that, you come up with hypotheses – what if you do something this way and not the other? And these hypotheses are often wrong. This is a lot harder and more creative than what specialists do at major IT companies, if I might say so.
Have you ever thought of working at a regular IT company?
I think that I can apply my knowledge both in research projects and IT. What I find most important is that projects that I work on would have an end user and be of use to people.
Why studying at a university is really useful

You’ve recently published an article on HABR which had a major response. Can you tell us what it was about?
I wanted to explain that the process of predicting the structure of two interacting molecules, a quite important thing when designing a drug, takes place with the help of computer modeling that’s based on comprehensible mathematical ideas. This is the subject matter of my article.
But there’s also an underlying message: I wanted to show how important it is to study, to get higher education that expands your outlook, and allows you to come up with unusual solutions. The knowledge that I got during my Bachelor’s and Master’s programs really have practical applications, sometimes in unobvious ways.
There’s a popular belief that if you work since your third year of Bachelor’s studies and already have an expertise in your field, there’s no need to enter a Master’s program. I have worked since my third year myself, and I see that a Master’s program offers new knowledge and helps expand one’s outlook nonetheless. For example, the algorithms that I currently use in my work are those that we discussed at lectures one year ago. At that time, I thought: “How boring, why do we even need to learn these things?”. And now I successfully apply these algorithms.

Can you give an example?
There’s quite a well-known fast Fourier transform algorithm, which is usually studied as part of a course on algorithms and data structures. Out of context, you only learn that it’s often applied by physicists, but you don’t understand what’s the use of it for a person who’s doing programming or bioinformatics. I couldn’t imagine I would ever need it. But when I started working on a docking algorithm, it turned out that this algorithm is almost completely based on the fast Fourier transform algorithm.
What feedback did you get for your article?
At first, I wanted it to be understandable both for people who don’t know much about biology and for those who don’t have lots of knowledge in maths and programming. It was really cool when people with no expertise in biology or math said that they understood everything. Some people on Habr wrote that even biologists got it (he laughs). When people say so, it’s very pleasant.
Do you plan to continue writing popular science articles?
At first, I didn’t. But now I think that I could continue doing that in collaboration with educational projects by JetBrains or Biocad. I think this may be a promising field.





