Malaria is what unites famous Italian poet Dante Alighieri, Portuguese traveller Vasco Da Gama, and Russian decembrist Alexander Odoevsky – it was their cause of death. This disease continues to affect many people even in our time. According to WHO, in 2019, there were 229 million cases of malaria registered globally with over 400,000 malaria deaths.
In 2019, malaria was found in 80 countries, including 108 cases in Russia. Most of all the disease affects those in the African continent – with over 90% of cases of malaria infection registered there.
It is a well-known fact that this disease is spread by mosquitoes. It is with their bites that Plasmodium parasites causing malaria get into the human body. A total of 400 species are considered to be malaria mosquitoes, but only 30 of them are the major vectors of the disease. However, until recently scientists had a reference genome of only one species.
“Such genomes are important for comparison of different species,” explains Anton Zamyatin, a researcher at ITMO University’s Laboratory of Genomic Diversity. “This way, we can see how far they are from each other in the classification, what makes them distinct, and which genome regions are responsible for this distinction. Thus, we know that some malaria mosquitoes feed on the blood of humans and other animals, while others bite humans only. It is important to pinpoint the genomic differences in such feeding behavior so that we can potentially affect the dietary preferences of certain species.”

What is a reference genome?
A team of researchers from Virginia Polytechnic Institute and State University (Virginia Tech), George Washington University, and ITMO University have launched a joint project which aims at assembling reference genomes of two species of African malaria mosquitoes: Anopheles coluzzii and Anopheles arabiensis
“Genomes can be assembled on various levels,” continues Anton Zamyatin. “During sequencing we get short sequences of symbols read from different regions of the genome. These sequences can be assembled into contigs, which are DNA segments. However, these segments don’t look the same way they would in a complete chromosome. It is just a collection of random information. The next level is scaffolding (orienting and sorting) of contigs in the same order that they appear in the actual genome. As soon as we have re-oriented the contigs and placed them in the right order, we can say that we have a genome at the level of scaffolds.”
This, however, is not the limit. These days, researchers can analyze genomes so deeply that the assemblies will resemble complete chromosomes. In other words, researchers can uncover the genome sequence in a chromosome from the beginning to the end.

The best and most precise assemblies are called reference genomes. They are stored in databases used all over the world as a standard. Such genomes help identify differences between individuals, species, and whole populations.
“Until recently, such a genome assembly existed only for a malaria mosquito Anopheles gambiae,” expounds Anton Zamyatin. “It is most likely to have been used for reference in studies of mosquito genomes. We wanted to assemble two more genomes so that ours are of equally high quality as the existing one. As a result, we created even better assemblies.”
Two years of work
The project started in 2018. At the time, genome separation and sequencing were already underway at Virginia Tech. There, researchers keep colonies of insects, separated by species, for experimental purposes. After that, ITMO University experts began assembling the genome.
“Assembling a mosquito’s genome is a moderately difficult task,” adds Anton Zamyatin. “On the one hand, its genome is quite large: there are about 300 million base pairs, which is only 10 times smaller than what humans have. On the other hand, mosquitoes only have three pairs of chromosomes, whereas humans have 23. One of the challenges here is that mosquito genomes are not sequenced individually. The genetic material taken from the entire colony, in other words, DNA from multiple individuals, was put into the sequencing machine at the same time. As a result, we can see individual features of mosquitoes that need to be neutralized in order to create a reference genome.”

That was not the only challenge. After the initial assembly, the researchers proceeded with scaffolding. For this process, they used the data about the spatial arrangement of chromatin inside a cell’s nucleus, which helps correctly orient contigs in relation to each other. For these purposes, the researchers used bioinformatics software that, however, did not always fulfil their needs. “At times we had to contact the developers and point out the mistakes we encountered,” remembers Anton Zamyatin. “Finally, we simply decided to write our own app. It is currently in development.”
Prospects
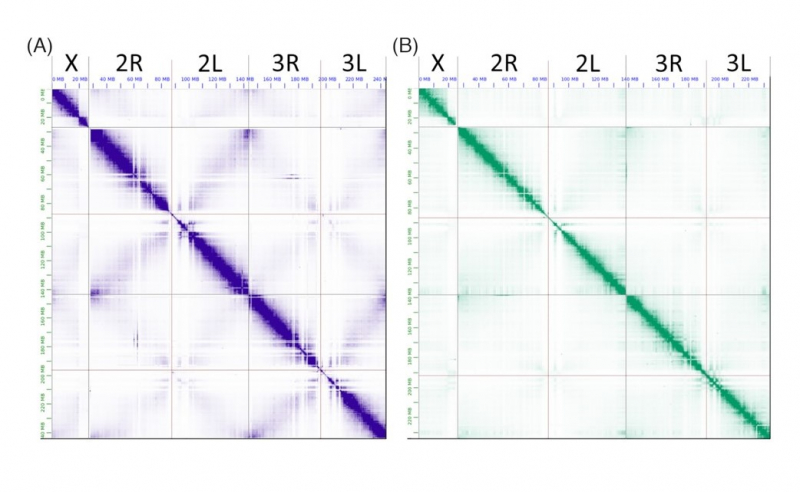
Apart from the assemblies of two genomes, the article offers a preliminary analysis of the acquired data. For instance, the researchers were looking for genome rearrangements – changes in the genome’s structure that occurred during evolution. These rearrangements can be responsible for the fact that species have different behavior and physiology.
“The sequence itself is fairly interesting, but then we proceed with functional annotation, where we identify regions of the genome that are responsible for genes present in a particular organism,” says Anton Zamyatin. “Over a species’ evolution, certain events can lead to changes in the sequence of genome regions, and we can see such changes in mosquitoes. It can affect their ability to transport malaria pathogens, their preferences for blood of a certain species, and so on.”
With this data, the scientists will be able to start new research projects aimed at preventing the spread of malaria. At the same time, ITMO University researchers are planning new initiatives related to assemblies of reference genomes of other species of malaria mosquitoes.
Reference: Anton Zamyatin, Pavel Avdeyev, Jiangtao Liang, Atashi Sharma, Chujia Chen, Varvara Lukyanchikova, Nikita Alexeev, Zhijian Tu, Max A Alekseyev, Igor V Sharakhov. Chromosome-level genome assemblies of the malaria vectors Anopheles coluzzii and Anopheles arabiensis. GigaScience, 2021/10.1093/gigascience/giab017