GWAS and its drawbacks
The method of genome-wide association studies (GWAS) made it possible to collect a considerable database of specific DNA variants associated with heightened risks of certain diseases. However, with GWAS it’s impossible to identify the particular genes that malfunction and cause a body-wide collapse. GWAS is limited by microchip-based genotyping that can only pinpoint a genome’s loci associated with a disease – and these loci can include dozens of genes with only one or two of them truly having to do with the illness. That’s why it’s hard to use GWAS when developing specific medical solutions.
All over the world, researchers are working to develop postprocessing methods for data acquired via GWAS. This is a rather challenging task because they have to process a large database with unknown contents – there is no way to know which associated genome loci to analyze and which to ignore.
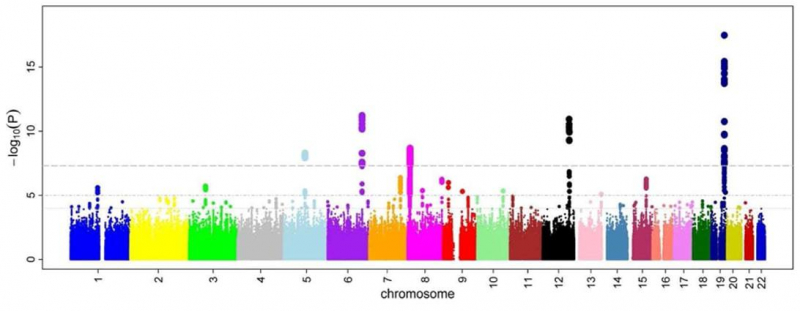
A Manhattan plot of some of the closely connected risk loci. Each point is a single-nucleotide polymorphism; its location is reflected on the X axis, while the Y axis shows the association level. The plot is taken from a GWAS study of microcirculatory disorders in blood vessels. Credit: M. Kamran Ikram, wikipedia.org (CC BY 2.5)
Scientists from ITMO’s Laboratory of Genomic Diversity with colleagues from the World-Class Research Centre for Personalized Medicine came up with their own solution to the problem. GPrior is a machine-learning-based tool that helps prioritize the genes connected to the heightened risk of a disease using positive unlabeled learning (where it’s possible to teach the model using only positive examples).
“In the majority of cases, we can safely say that one particular gene isn’t responsible for the phenotype’s development. The data acquired via GWAS leaves us with a limited number of positive examples of genes whose role in the phenotype’s development we can securely identify, and a great number of genes, of which we know nothing for certain – they can be either positive or negative. Our algorithm takes it upon itself to solve this riddle and mark all of these undetermined cases,” explains Nikita Kolosov, author of the study, a researcher at the International Laboratory "Computer Technologies” and programmer at the Laboratory of Genomic Diversity and the World-Class Research Centre for Personalized Medicine.
Separate the wheat from the chaff
How can we train an algorithm to see what’s important in a large array of data? The classical approach to training classifiers, such as algorithms for detecting tumors in images, relies on a great amount of both positive (images with tumors in them) and negative (images that definitely don’t display one) examples. This method isn’t applicable in gene prioritization – there is simply too much unidentified and unmarked data in the input.
That’s why the researchers decided to take another approach: they employ five different classifiers that consecutively analyze a data set assigning a functional annotation to each gene in it. The algorithm uses two types of features that the authors refer to as SNP (single-nucleotide polymorphism) and gene levels. Similar nucleotide chains on homologous chromosome loci are analyzed at the SNP level, while at the gene level functional annotations are assigned, as the name suggests, to specific genes. This way, it’s possible to create a table with all genes and their functions.
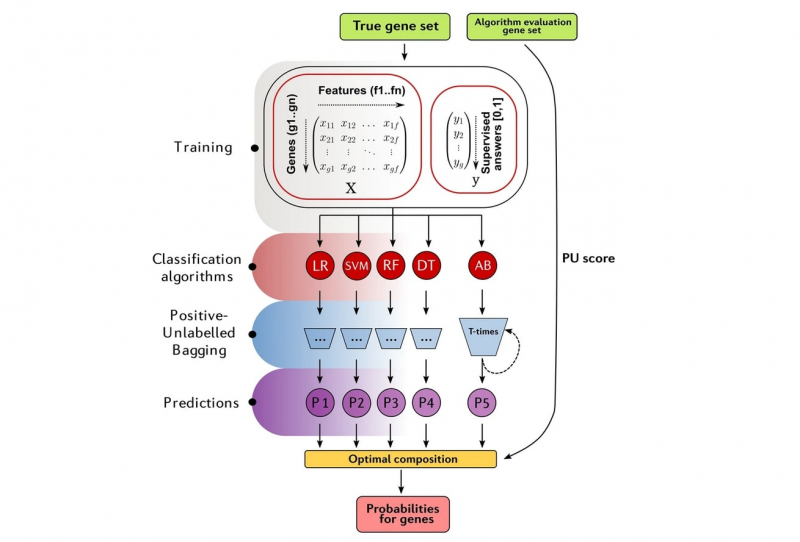
GPrior ensemble positive-unlabeled learning framework. Credit: nature.org
“We take the GWAS output and the gene variants that correlate to a particular phenotype (such as that of a disease) and link them to specific genes by assigning them various functional annotations. We also use additional annotations such as expression level in different tissues (kidneys, liver, etc.) and then use these to prioritize all the genes that we analyze. As a result, we get a ranked list of genes in order of their likelihood to be connected to a particular phenotype’s development,” explains Nikita Kolosov.
An all-purpose ensemble
According to the authors of the study, the new method is not only highly efficient but also is a flexible multi-purpose tool. The algorithm makes it possible to take an individual approach to each new dataset thanks to the ensemble of five classifiers at its core that consecutively analyze a large amount of data.
The new algorithm is available on GitHub and open to researchers from all over the world and the developers are planning to constantly support and further improve it.
In general, though, the authors admit that there is yet no satisfactory solution for identifying risk genes in polygenic diseases. For now, the task is still complicated by a great number of problems.

Nikita Kolosov. Photo by Ekaterina Shevyreva, ITMO.NEWS
Nikita Kolosov maintains that the very step from GWAS to specific genetic elements potentially responsible for the development of a disease is a rather non-trivial one. At the same time, it is a task that has to be solved because once we discover a disease’s genetic causes, it will be possible to develop efficient pharmaceutical treatment.
“Understanding the genetic structure and identifying the combination of genes behind a particular disease would amount to a breakthrough discovery,” says the researcher. “Many polygenic conditions, such as schizophrenia or coronary heart disease, can be attributed to a joint effect of mutations in a great number of genes interacting with each other. Deconstructing such complex features and identifying their genetic reasons is an important task for medicine. And collaborations between leading genomic and bioinformatic centers, such as ITMO, World-Class Research Centre for Personalized Medicine, and Broad Institute in our case, is the key for new effective solutions.”
Reference: Nikita Kolosov, Mark J. Daly, Mykyta Artomov. Prioritization of disease genes from GWAS using ensemble based positive-unlabeled learning. European Journal of Human Genetics, 29, 1527–1535 (2021).





