Why do we need new storage methods?
There are many ways to store information in the digital form. Data is kept on HDDs, SSDs, flash drives, memory cards, optical media (CD, DVD, Blu-Ray), and in cloud storage. But each technology has its drawbacks.
First of all, no hard drive today exceeds 20 terabytes in volume. In the meantime, the International Data Corporation and Seagate predict that the global volume of data will reach 175-180 zetabytes by 2025. This translates to approximately one billion terabytes.
Secondly, the longevity of hard drives depends on their storage conditions – for instance, it’s essential to maintain a certain level of temperature and humidity. Because of this, data storage centers will have to spend even more energy and, as a result, produce a larger environmental footprint.
Lastly, data stored in this manner can be hacked and stolen. Because of all these reasons, chemists and biologists around the world have been working on new ways to store data for a long time and with the least resources.
See also:
So, what are scientists proposing?
Data and DNA
In most cases, before data is recorded onto conventional media – such as a hard drive – it must be encrypted and represented as a chain of zeros and ones. DNA-based data storage relies on the same principle, but converts every chain into a sequence of nucleotide bases – adenine, cytosine, guanine, and thymine – which form DNA base pairs. As a result, the data becomes a strand of synthetic DNA. In order to be read, it must be sequenced (a process that involves identifying the order of nucleotides in a DNA molecule) and decoded.
The startup Catalog has already succeeded in testing this concept by converting the entirety of Wikipedia into DNA. Now, the company is working on making it possible to quickly find information among large swaths of data. In the future, this would help accomplish numerous tasks, such as identifying fraudulent transactions or discovering new fossil fuel deposits.
Data and macromolecules
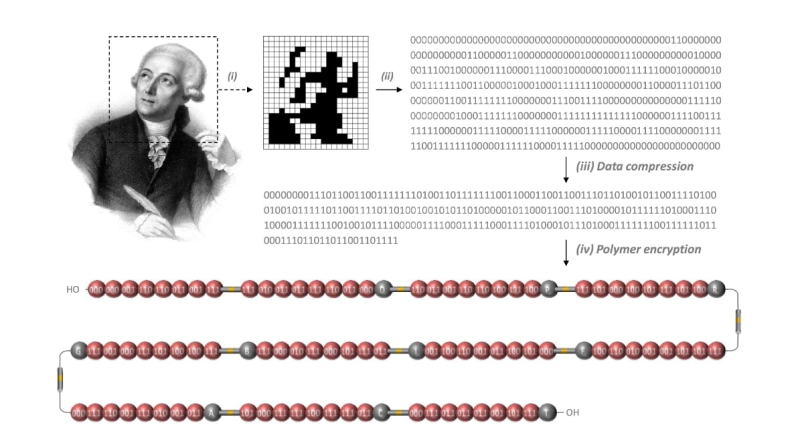
An image illustrating the encryption of Antoine de Lavoisier's portrait onto a single macromolecure. Credit: comptes-rendus.academie-sciences.fr
In the meantime, a research group has succeeded in storing a portrait of the French naturalist and chemist Antoine de Lavoisier inside a single macromolecule. First, they divided the image into 440 pixels, which they converted into 440 bits of binary code. The code was then compressed into a 264-bit string and translated into a chemical sequence of eight monomers containing the encoded portrait, as well as one decomposable spacer and ten mass spectrometry tags. The tags facilitate the macromolecule’s decoding with mass spectrometry.
Data and mixtures of organic molecules
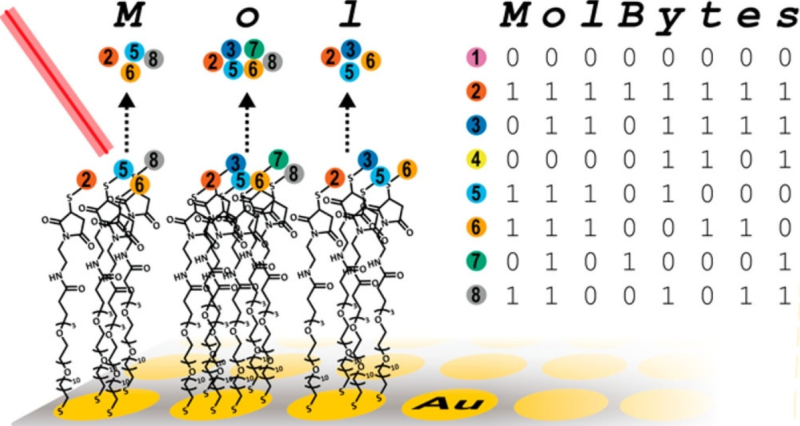
A schematic of recording information using a mixture of organic molecules. Credit: pubs.acs.org
The group headed by George M. Whitesides successfully used organic rather than synthetic molecules to store an image of the mathematician Claude Shannon, an image of the woodblock print The Great Wave off Kanagawa by Hokusai, and the text of Richard Feynman’s lecture There's Plenty of Room at the Bottom.
In their experiment, the researchers used 32 oligopeptides with various molecular mass. The information was first encoded in binary, and then those of the eight bits in a byte that had the value 1 were assigned an oligopeptide. After that, the encoded molecular information (400 kilobits) was transferred to an array plate, where it spread over 1,536 microwells, which help identify a bit’s location when the information is decoded using mass spectrometry. It took the researchers 20 hours to record and then read their message (the writing speed being 8 bit/s, and the reading – 20 bit/s).
Data and mixtures of fluorescent molecules
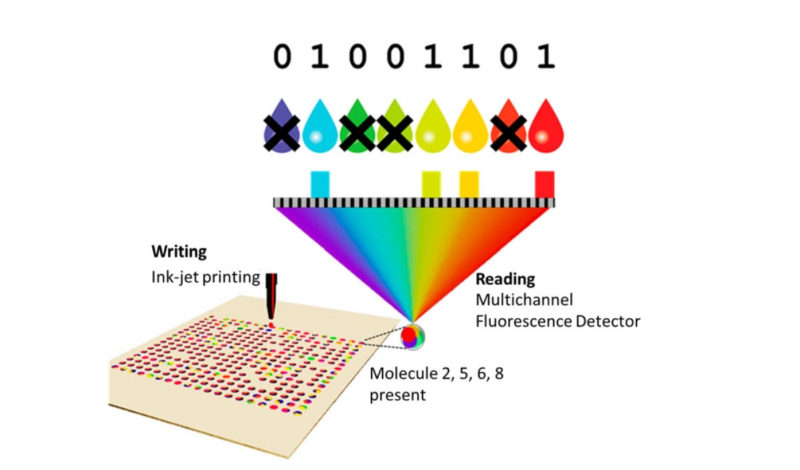
A schematic of the process of recording and reading information using fluorescent molecules. Credit: ncbi.nlm.nih.gov
George M. Whitesides’ group is working on a faster storage method, as well: the researchers were able to record about 400 kilobits at the speed of 128 bit/s and read it at a rate of 469 bit/s. This achievement was made possible by inkjet printing, molecules with fluorescent dyes, and a microscope equipped with a multichannel fluorescence detector. The encoding procedure itself is similar to that described above – the information is encoded in binary, translated to molecular, and transferred to arrays with microwells organized into microsquares. However, whereas a distinguishing feature of organic molecules is their mass, this time the fluorescent dyes helped tell the molecules apart, which significantly accelerated the reading process. In this experiment, the researchers used seven colors to encode an image of the physicist Michael Faraday and the first part of his work Experimental Researches in Electricity. In the future, it will be possible to increase the number of colors and the amount of recorded information.
In 2022, George M. Whitesides took part in the Conference on Robotization of Chemical Technologies, organized by ITMO’s Infochemistry Scientific Center. There, scientists, students, and representatives of tech companies discussed the topical issues of robotization in chemistry, such as how robots are used to develop smart functional materials, collect databases, establish new research fields, and more.






